The broom.helpers package offers a suite of functions
that make easy to interact, add information, and manipulate tibbles
created with broom::tidy() (and friends).
The suite includes functions to group regression model terms by variable, insert reference and header rows for categorical variables, add variable labels, and more.
As a motivating example, let’s summarize a logistic regression model with a forest plot and in a table.
To begin, let’s load our packages.
library(broom.helpers)
library(gtsummary)
library(ggplot2)
library(dplyr)
# paged_table() was introduced only in rmarkdwon v1.2
print_table <- function(tab) {
if (packageVersion("rmarkdown") >= "1.2") {
rmarkdown::paged_table(tab)
} else {
knitr::kable(tab)
}
}Our model predicts tumor response using chemotherapy treatment and tumor grade. The data set we’re utilizing has already labelled the columns using the labelled package. The column labels will be carried through to our figure and table.
model_logit <- glm(response ~ trt + grade, trial, family = binomial)
broom::tidy(model_logit)
#> # A tibble: 4 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) -0.879 0.305 -2.88 0.00400
#> 2 trtDrug B 0.194 0.311 0.625 0.532
#> 3 gradeII -0.0647 0.381 -0.170 0.865
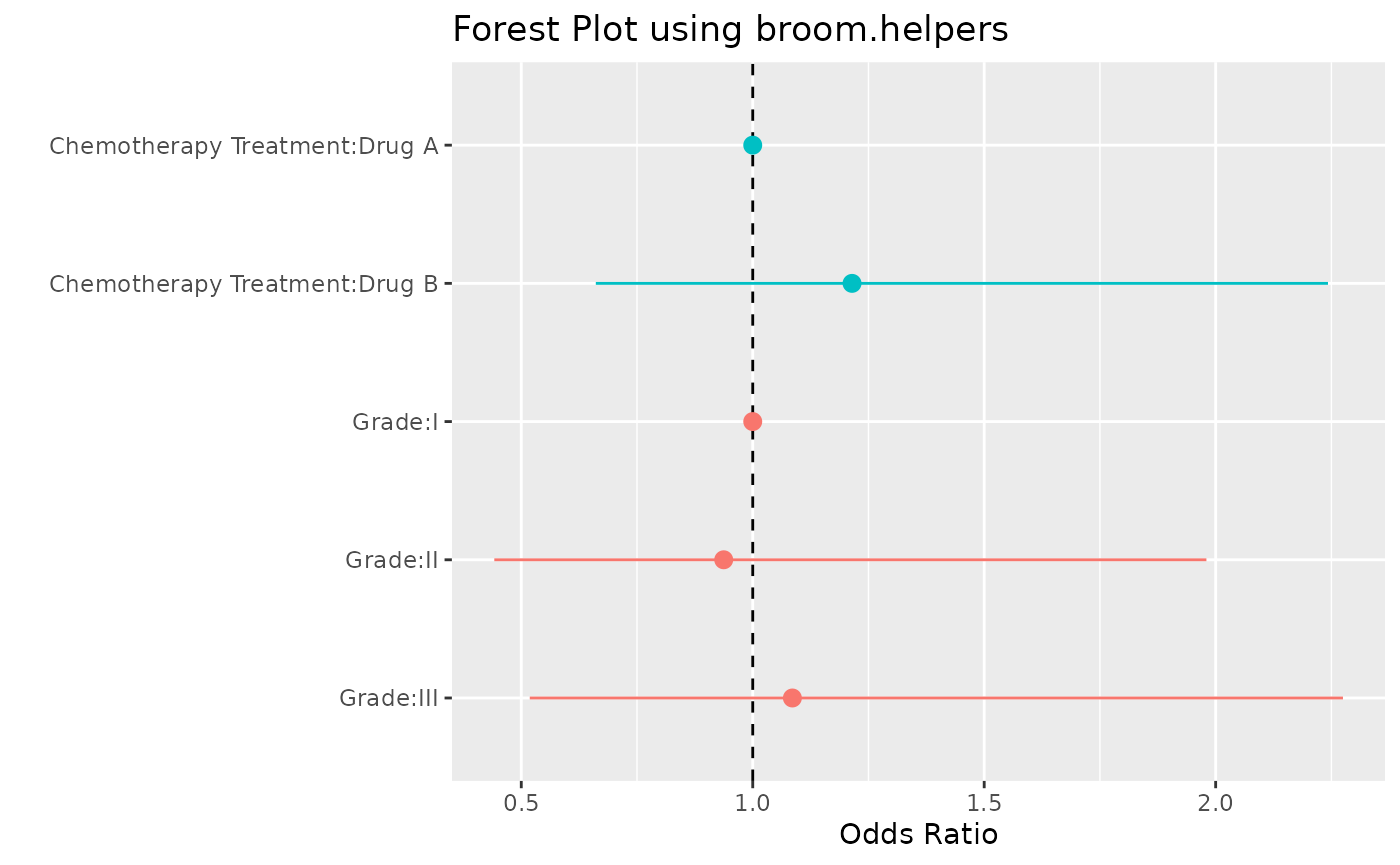
#> 4 gradeIII 0.0822 0.376 0.219 0.827Forest Plot
To create the figure, we’ll need to add some information to the tidy
tibble, i.e. we’ll need to group the terms that belong to the same
variable, add the reference row, etc. Parsing this information can be
difficult, but the broom.helper package has made it
simple.
tidy_forest <-
model_logit |>
# perform initial tidying of the model
tidy_and_attach(exponentiate = TRUE, conf.int = TRUE) |>
# adding in the reference row for categorical variables
tidy_add_reference_rows() |>
# adding a reference value to appear in plot
tidy_add_estimate_to_reference_rows() |>
# adding the variable labels
tidy_add_term_labels() |>
# removing intercept estimate from model
tidy_remove_intercept()
tidy_forest
#> # A tibble: 5 × 16
#> term variable var_label var_class var_type var_nlevels contrasts
#> <chr> <chr> <chr> <chr> <chr> <int> <chr>
#> 1 trtDrug A trt Chemotherapy Trea… character dichoto… 2 contr.tr…
#> 2 trtDrug B trt Chemotherapy Trea… character dichoto… 2 contr.tr…
#> 3 gradeI grade Grade factor categor… 3 contr.tr…
#> 4 gradeII grade Grade factor categor… 3 contr.tr…
#> 5 gradeIII grade Grade factor categor… 3 contr.tr…
#> # ℹ 9 more variables: contrasts_type <chr>, reference_row <lgl>, label <chr>,
#> # estimate <dbl>, std.error <dbl>, statistic <dbl>, p.value <dbl>,
#> # conf.low <dbl>, conf.high <dbl>Note: we used tidy_and_attach() instead
of broom::tidy(). broom.helpers functions
needs a copy of the original model. To avoid passing the model at each
step, the easier way is to attach the model as an attribute of the
tibble with tidy_attach_model().
tidy_and_attach() is simply a shortcut of
model |> broom::tidy() |> tidy_and_attach(model).
We now have a tibble with every piece of information we need to
create our forest plot using ggplot2.
tidy_forest |>
mutate(
plot_label = paste(var_label, label, sep = ":") |>
forcats::fct_inorder() |>
forcats::fct_rev()
) |>
ggplot(aes(x = plot_label, y = estimate, ymin = conf.low, ymax = conf.high, color = variable)) +
geom_hline(yintercept = 1, linetype = 2) +
geom_pointrange() +
coord_flip() +
theme(legend.position = "none") +
labs(
y = "Odds Ratio",
x = " ",
title = "Forest Plot using broom.helpers"
)
Note:: for more advanced and nicely formatted plots
of model coefficients, look at ggstats::ggcoef_model() and
its dedicated
vignette. ggstats::ggcoef_model() internally uses
broom.helpers.
Table Summary
In addition to aiding in figure creation, the broom.helpers package can help summarize a model in a table. In the example below, we add header and reference rows, and utilize existing variable labels. Let’s change the labels shown in our summary table as well.
tidy_table <-
model_logit |>
# perform initial tidying of the model
tidy_and_attach(exponentiate = TRUE, conf.int = TRUE) |>
# adding in the reference row for categorical variables
tidy_add_reference_rows() |>
# adding the variable labels
tidy_add_term_labels() |>
# add header row
tidy_add_header_rows() |>
# removing intercept estimate from model
tidy_remove_intercept()
# print summary table
options(knitr.kable.NA = "")
tidy_table |>
# format model estimates
select(label, estimate, conf.low, conf.high, p.value) |>
mutate(across(all_of(c("estimate", "conf.low", "conf.high")), style_ratio)) |>
mutate(across(p.value, style_pvalue)) |>
print_table()Note:: for more advanced and nicely formatted tables
of model coefficients, look at gtsummary::tbl_regression()
and its dedicated
vignette. gtsummary::tbl_regression() internally uses
broom.helpers.
All-in-one function
There is also a handy wrapper, called tidy_plus_plus(),
for the most commonly used tidy_*() functions, and they can
be executed with a single line of code:
model_logit |>
tidy_plus_plus(exponentiate = TRUE)
#> # A tibble: 5 × 18
#> term variable var_label var_class var_type var_nlevels contrasts
#> <chr> <chr> <chr> <chr> <chr> <int> <chr>
#> 1 trtDrug A trt Chemotherapy Trea… character dichoto… 2 contr.tr…
#> 2 trtDrug B trt Chemotherapy Trea… character dichoto… 2 contr.tr…
#> 3 gradeI grade Grade factor categor… 3 contr.tr…
#> 4 gradeII grade Grade factor categor… 3 contr.tr…
#> 5 gradeIII grade Grade factor categor… 3 contr.tr…
#> # ℹ 11 more variables: contrasts_type <chr>, reference_row <lgl>, label <chr>,
#> # n_obs <dbl>, n_event <dbl>, estimate <dbl>, std.error <dbl>,
#> # statistic <dbl>, p.value <dbl>, conf.low <dbl>, conf.high <dbl>
model_logit |>
tidy_plus_plus(exponentiate = TRUE) |>
print_table()See the documentation of tidy_plus_plus() for the full
list of available options.
Advanced examples
broom.helpers can also handle different contrasts for
categorical variables and the use of polynomial terms for continuous
variables.
Polynomial terms
When polynomial terms of a continuous variable are defined with
stats::poly(), broom.helpers will be able to
identify the corresponding variable, create appropriate labels and add
header rows.
model_poly <- glm(response ~ poly(age, 3) + ttdeath, na.omit(trial), family = binomial)
model_poly |>
tidy_plus_plus(
exponentiate = TRUE,
add_header_rows = TRUE,
variable_labels = c(age = "Age in years")
) |>
print_table()Different type of contrasts
By default, categorical variables are coded with a treatment
contrasts (see stats::contr.treatment()). With such
contrasts, model coefficients correspond to the effect of a modality
compared with the reference modality (by default, the first one).
tidy_add_reference_rows() allows to add a row for this
reference modality and
tidy_add_estimate_to_reference_rows() will populate the
estimate value of these references rows by 0 (or 1 if
exponentiate = TRUE). tidy_add_term_labels()
is able to retrieve the label of the factor level associated with a
specific model term.
model_1 <- glm(
response ~ stage + grade * trt,
gtsummary::trial,
family = binomial
)
model_1 |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_reference_rows() |>
tidy_add_estimate_to_reference_rows(exponentiate = TRUE) |>
tidy_add_term_labels() |>
print_table()Using stats::contr.treatment(), it is possible to
defined alternative reference rows. It will be properly managed by
broom.helpers.
model_2 <- glm(
response ~ stage + grade * trt,
gtsummary::trial,
family = binomial,
contrasts = list(
stage = contr.treatment(4, base = 3),
grade = contr.treatment(3, base = 2),
trt = contr.treatment(2, base = 2)
)
)
model_2 |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_reference_rows() |>
tidy_add_estimate_to_reference_rows(exponentiate = TRUE) |>
tidy_add_term_labels() |>
print_table()You can also use sum contrasts (cf. stats::contr.sum()).
In that case, each model coefficient corresponds to the difference of
that modality with the grand mean. A variable with 4 modalities will be
coded with 3 terms. However, a value could be computed (using
emmeans::emmeans()) for the last modality, corresponding to
the difference of that modality with the grand mean and equal to sum of
all other coefficients multiplied by -1. broom.helpers will
identify categorical variables coded with sum contrasts and could
retrieve an estimate value for the reference term.
model_3 <- glm(
response ~ stage + grade * trt,
gtsummary::trial,
family = binomial,
contrasts = list(
stage = contr.sum,
grade = contr.sum,
trt = contr.sum
)
)
model_3 |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_reference_rows() |>
tidy_add_estimate_to_reference_rows(exponentiate = TRUE) |>
tidy_add_term_labels() |>
print_table()Other types of contrasts exist, like Helmert
(contr.helmert()) or polynomial
(contr.poly()). They are more complex as a modality will be
coded with a combination of terms. Therefore, for such contrasts, it
will not be possible to associate a specific model term with a level of
the original factor. broom.helpers will not add a reference
term in such case.
model_4 <- glm(
response ~ stage + grade * trt,
gtsummary::trial,
family = binomial,
contrasts = list(
stage = contr.poly,
grade = contr.helmert,
trt = contr.poly
)
)
model_4 |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_reference_rows() |>
tidy_add_estimate_to_reference_rows(exponentiate = TRUE) |>
tidy_add_term_labels() |>
print_table()Pairwise contrasts of categorical variable
Pairwise contrasts of categorical variables could be computed with
tidy_add_pairwise_contrasts().
model_logit <- glm(response ~ age + trt + grade, trial, family = binomial)
model_logit |>
tidy_and_attach() |>
tidy_add_pairwise_contrasts() |>
print_table()
model_logit |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_pairwise_contrasts() |>
print_table()
model_logit |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_pairwise_contrasts(pairwise_reverse = FALSE) |>
print_table()
model_logit |>
tidy_and_attach(exponentiate = TRUE) |>
tidy_add_pairwise_contrasts(keep_model_terms = TRUE) |>
print_table()Column Details
Below is a summary of the additional columns that may be added by a
broom.helpers function. The table includes the column name,
the function that adds the column, and a short description of the
information in the column.
| Column | Function | Description |
|---|---|---|
| contrasts | tidy_add_contrasts() |
Contrasts used for categorical variables. Require “variable” column. If needed, will automatically apply tidy_identify_variables(). |
| contrasts_type | tidy_add_contrasts() |
Type of contrasts (“treatment”, “sum”, “poly”, “helmert”, “sdif”, “other” or “no.contrast”). “pairwise is used for pairwise contrasts computed with tidy_add_pairwise_contrasts(). |
| exposure | tidy_add_n() |
Exposure time (for Poisson and Cox models) |
| group_by | tidy_group_by() |
Grouping variable (particularly for multinomial or multi-components models). |
| header_row | tidy_add_header_rows() |
Logical indicating if a row is a header row for variables with several terms. Is equal to NA for variables who do not have an header row.Require “label” column. If needed, will automatically apply tidy_add_term_labels().It is better to apply tidy_add_header_rows() after other tidy_* functions |
| instrumental | tidy_identify_variables() |
For “fixest” models, indicate if a variable was instrumental. |
| label | tidy_add_term_labels() |
String of term labels based on (1) labels provided in labels argument if provided; (2) factor levels for categorical variables coded with treatment, SAS or sum contrasts; (3) variable labels when there is only one term per variable; and (4) term name otherwise.Require “variable_label” column. If needed, will automatically apply tidy_add_variable_labels().Require “contrasts” column. If needed, will automatically apply tidy_add_contrasts(). |
| n_event | tidy_add_n() |
Number of events (for binomial and multinomial logistic models, Poisson and Cox models) |
| n_ind | tidy_add_n() |
Number of individuals (for Cox models) |
| n_obs | tidy_add_n() |
Number of observations |
| original_term | tidy_disambiguate_terms(), tidy_multgee(), tidy_zeroinfl() or tidy_identify_variables() |
Original term before disambiguation. This columns is added only when disambiguation is needed (i.e. for mixed models). Also used for “multgee”, “zeroinfl” and “hurdle” models. For instrumental variables in “fixest” models, the “fit_” prefix is removed, and the original terms is stored in this column. |
| reference_row | tidy_add_reference_rows() |
Logical indicating if a row is a reference row for categorical variables using a treatment or a sum contrast. Is equal to NA for variables who do not have a reference row.Require “contrasts” column. If needed, will automatically apply tidy_add_contrasts().tidy_add_reference_rows() will not populate the label of the reference term. It is therefore better to apply tidy_add_term_labels() after tidy_add_reference_rows() rather than before. |
| var_class | tidy_identify_variables() |
Class of the variable. |
| var_label | tidy_add_variable_labels() |
String of variable labels from the model. Columns labelled with the labelled package are retained. It is possible to pass a custom label for an interaction term with the labels argument. Require “variable” column. If needed, will automatically apply tidy_identify_variables(). |
| var_nlevels | tidy_identify_variables() |
Number of original levels for categorical variables |
| var_type | tidy_identify_variables() |
One of “intercept”, “continuous”, “dichotomous”, “categorical”, “interaction”, “ran_pars” or “ran_vals” |
| variable | tidy_identify_variables() |
String of variable names from the model. For categorical variables and polynomial terms defined with stats::poly(), terms belonging to the variable are identified. |
Note: tidy_add_estimate_to_reference_rows() does not
create an additional column; rather, it populates the ‘estimate’ column
for reference rows.
Additional attributes
Below is a list of additional attributes that
broom.helpers may attached to the results. The table
includes the attribute name, the function that adds the attribute, and a
short description.
| Attribute | Function | Description |
|---|---|---|
| coefficients_label | tidy_add_coefficients_type() |
Coefficients label |
| coefficients_type | tidy_add_coefficients_type() |
Type of coefficients |
| component | tidy_zeroinfl() |
component argument passed to tidy_zeroinfl() |
| conf.level | tidy_and_attach() |
Level of confidence used for confidence intervals |
| exponentiate | tidy_and_attach() |
Indicates if estimates were exponentiated |
| Exposure | tidy_add_n() |
Total of exposure time |
| N_event | tidy_add_n() |
Total number of events |
| N_ind | tidy_add_n() |
Total number of individuals (for Cox models) |
| N_obs | tidy_add_n() |
Total number of observations |
| term_labels | tidy_add_term_labels() |
Custom term labels passed to tidy_add_term_labels() |
| variable_labels | tidy_add_variable_labels() |
Custom variable labels passed to tidy_add_variable_labels() |
Supported models
| Model | Notes |
|---|---|
betareg::betareg() |
Use tidy_parameters() as tidy_fun with component argument to control with coefficients to return. broom::tidy() does not support the exponentiate argument for betareg models, use tidy_parameters() instead. |
biglm::bigglm() |
|
brms::brm() |
broom.mixed package required |
cmprsk::crr() |
Limited support. It is recommended to use tidycmprsk::crr() instead. |
fixest::feglm() |
May fail with R <= 4.0. |
fixest::femlm() |
May fail with R <= 4.0. |
fixest::feNmlm() |
May fail with R <= 4.0. |
fixest::feols() |
May fail with R <= 4.0. |
gam::gam() |
|
geepack::geeglm() |
|
glmmTMB::glmmTMB() |
broom.mixed package required |
glmtoolbox::glmgee() |
|
lavaan::lavaan() |
Limited support for categorical variables |
lfe::felm() |
|
lme4::glmer.nb() |
broom.mixed package required |
lme4::glmer() |
broom.mixed package required |
lme4::lmer() |
broom.mixed package required |
logitr::logitr() |
Requires logitr >= 0.8.0 |
MASS::glm.nb() |
|
MASS::polr() |
|
mgcv::gam() |
Use default tidier broom::tidy() for smooth terms only, or gtsummary::tidy_gam() to include parametric terms |
mice::mira |
Limited support. If mod is a mira object, use tidy_fun = function(x, ...) {mice::pool(x) |> mice::tidy(...)} |
mmrm::mmrm() |
|
multgee::nomLORgee() |
Experimental support. Use tidy_multgee() as tidy_fun. |
multgee::ordLORgee() |
Experimental support. Use tidy_multgee() as tidy_fun. |
nnet::multinom() |
|
ordinal::clm() |
Limited support for models with nominal predictors. |
ordinal::clmm() |
Limited support for models with nominal predictors. |
parsnip::model_fit |
Supported as long as the type of model and the engine is supported. |
plm::plm() |
|
pscl::hurdle() |
Use tidy_zeroinfl() as tidy_fun. |
pscl::zeroinfl() |
Use tidy_zeroinfl() as tidy_fun. |
rstanarm::stan_glm() |
broom.mixed package required |
stats::aov() |
Reference rows are not relevant for such models. |
stats::glm() |
|
stats::lm() |
|
stats::nls() |
Limited support |
survey::svycoxph() |
|
survey::svyglm() |
|
survey::svyolr() |
|
survival::cch() |
`Experimental support. |
survival::clogit() |
|
survival::coxph() |
|
survival::survreg() |
|
tidycmprsk::crr() |
|
VGAM::vglm() |
Limited support. It is recommended to use tidy_parameters() as tidy_fun. |
Note: this list of models has been tested. broom.helpers
may or may not work properly or partially with other types of models. Do
not hesitate to provide feedback on GitHub.